Population Dynamics#
Teng-Jui Lin
Content adapted from UW AMATH 301, Beginning Scientific Computing, in Spring 2020.
Phase portraits
Population dynamics models
Predator-prey model
Competition model
Predator-pray model#
Let
where
Problem Statement. With the initial conditions of
Base case of
Changing
Changing
Changing
Changing
in
Base case#
import numpy as np
import matplotlib.pyplot as plt
import scipy
from scipy import integrate
# model params
a = 2
b = 1
c = 1
d = 1
# initial conditions
x0 = 1
y0 = 1
initial_val = np.array([x0, y0])
# time array
t_initial = 0
t_final = 12*2
t = np.linspace(t_initial, t_final, 1000)
# ode system
dxdt = lambda x, y : a*x - b*x*y
dydt = lambda x, y : -c*y + d*x*y
ode_syst = lambda t, z : np.array([dxdt(*z), dydt(*z)])
# ode soln
ode_soln = scipy.integrate.solve_ivp(ode_syst, [t_initial, t_final], initial_val, t_eval=t)
# quiver grid
xvec = np.linspace(0, 15, 20)
yvec = np.linspace(0, 15, 20)
X, Y = np.meshgrid(xvec, yvec)
# plot settings
%config InlineBackend.figure_format = 'retina'
%matplotlib inline
plt.rcParams.update({
'font.family': 'Arial', # Times New Roman, Calibri
'font.weight': 'normal',
'mathtext.fontset': 'cm',
'font.size': 18,
'lines.linewidth': 2,
'axes.linewidth': 2,
'axes.spines.top': False,
'axes.spines.right': False,
'axes.titleweight': 'bold',
'axes.titlesize': 18,
'axes.labelweight': 'bold',
'xtick.major.size': 8,
'xtick.major.width': 2,
'ytick.major.size': 8,
'ytick.major.width': 2,
'figure.dpi': 80,
'legend.framealpha': 1,
'legend.edgecolor': 'black',
'legend.fancybox': False,
'legend.fontsize': 14
})
fig, ax = plt.subplots(figsize=(4, 4))
# phase portrait
scale = np.sqrt(dxdt(X, Y)**2 + dydt(X, Y)**2)
ax.quiver(X, Y, dxdt(X, Y)/scale, dydt(X, Y)/scale, scale, cmap='winter_r', scale=20, width=0.005) # regular
ax.plot(*initial_val, 'o', color='black', label='Initial')
ax.plot(ode_soln.y[0], ode_soln.y[1], color='black', zorder=0.5)
ax.plot(ode_soln.y[0, -1], ode_soln.y[1, -1], 'X', color='red', zorder=4, label='Final')
# plot settings
ax.set_xlim(0, 15)
ax.set_ylim(0, 15)
ax.set_xlabel('Rabbits')
ax.set_ylabel('Foxes')
ax.set_title(f'Phase Portrait')
ax.set_aspect('equal')
ax.legend(loc='upper right')
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:4: RuntimeWarning: invalid value encountered in true_divide
after removing the cwd from sys.path.
<matplotlib.legend.Legend at 0x19ae3abccc8>
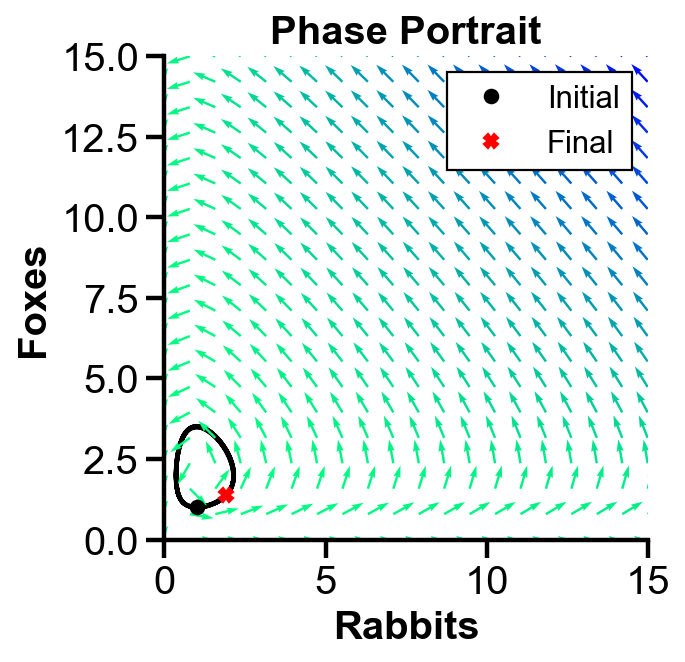
▲ The figure above shows the phase portrait of the rabbit-fox model with parameters
fig, ax = plt.subplots(figsize=(5, 4))
ax.plot(t, ode_soln.y[0], label='Rabbits')
ax.plot(t, ode_soln.y[1], '--', label='Foxes')
# plot settings
ax.set_xlim(0)
ax.set_ylim(0)
ax.set_xlabel('$t$')
ax.set_ylabel('Population')
ax.set_title(f'Population over time')
ax.legend(loc='upper left', bbox_to_anchor=(1.05, 1.05))
<matplotlib.legend.Legend at 0x19ae5b888c8>
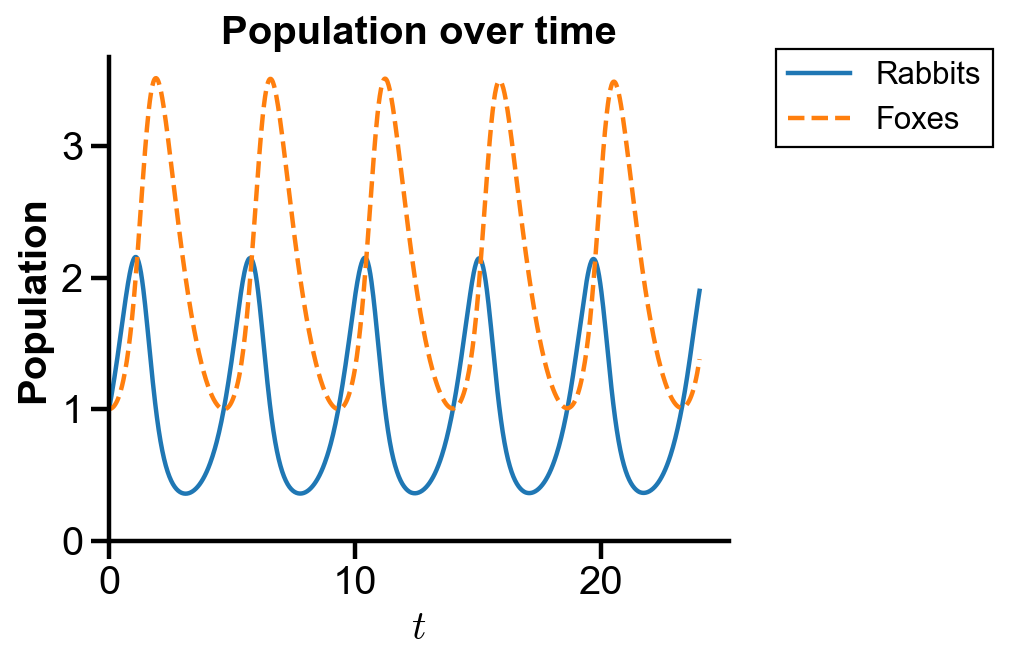
▲ The figure above shows the population of rabbits and foxes with parameters
Changing
# model params
a = np.arange(0, 5, 0.5)
b = 1
c = 1
d = 1
# initial conditions
x0 = 1
y0 = 1
initial_val = np.array([x0, y0])
# time array
t_initial = 0
t_final = 12*2
t = np.linspace(t_initial, t_final, 1000)
# quiver grid
xvec = np.linspace(0, 15, 15)
yvec = np.linspace(0, 15, 15)
X, Y = np.meshgrid(xvec, yvec)
row = 2
col = 5
fig1, axs1 = plt.subplots(row, col, figsize=(col*3, row*3), sharex=True, sharey=True)
fig1.add_subplot(111, frameon=False)
plt.tick_params(labelcolor='none', which='both', top=False, bottom=False, left=False, right=False) # hide tick and tick label of the big axis
plt.xlabel('Rabbits')
plt.ylabel('Foxes')
plt.title('Phase Portrait', y=1.1)
fig2, axs2 = plt.subplots(row, col, figsize=(col*3, row*3-1), sharex=True, sharey=True)
fig2.add_subplot(111, frameon=False)
plt.tick_params(labelcolor='none', which='both', top=False, bottom=False, left=False, right=False) # hide tick and tick label of the big axis
plt.xlabel('$t$')
plt.ylabel('Population')
plt.title('Population over time', y=1.1)
for row_i in range(row):
for col_i in range(col):
i = row_i*col + col_i
# ode system
dxdt = lambda x, y : a[i]*x - b*x*y
dydt = lambda x, y : -c*y + d*x*y
ode_syst = lambda t, z : np.array([dxdt(*z), dydt(*z)])
# ode soln
ode_soln = scipy.integrate.solve_ivp(ode_syst, [t_initial, t_final], initial_val, t_eval=t)
# phase portrait
scale = np.sqrt(dxdt(X, Y)**2 + dydt(X, Y)**2)
axs1[row_i, col_i].quiver(X, Y, dxdt(X, Y)/scale, dydt(X, Y)/scale, scale, cmap='winter_r', scale=15, width=0.008) # regular
axs1[row_i, col_i].plot(*initial_val, 'o', color='black', label='Initial')
axs1[row_i, col_i].plot(ode_soln.y[0], ode_soln.y[1], color='black', zorder=0.5)
axs1[row_i, col_i].plot(ode_soln.y[0, -1], ode_soln.y[1, -1], 'X', color='red', zorder=4, label='Final')
# plot settings
axs1[row_i, col_i].set_xlim(0, 15)
axs1[row_i, col_i].set_ylim(0, 15)
axs1[row_i, col_i].set_title(f'a = {a[i] :.1f}')
axs1[row_i, col_i].set_aspect('equal')
axs2[row_i, col_i].plot(t, ode_soln.y[0], label='Rabbits')
axs2[row_i, col_i].plot(t, ode_soln.y[1], label='Foxes')
axs2[row_i, col_i].set_xlim(t_initial, t_final)
axs2[row_i, col_i].set_ylim(0, 15)
axs2[row_i, col_i].set_title(f'a = {a[i] :.1f}')
# compromised legend for common xylabels
if row_i == 0 and col_i == (col-1):
axs1[row_i, col_i].legend()
axs2[row_i, col_i].legend()
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
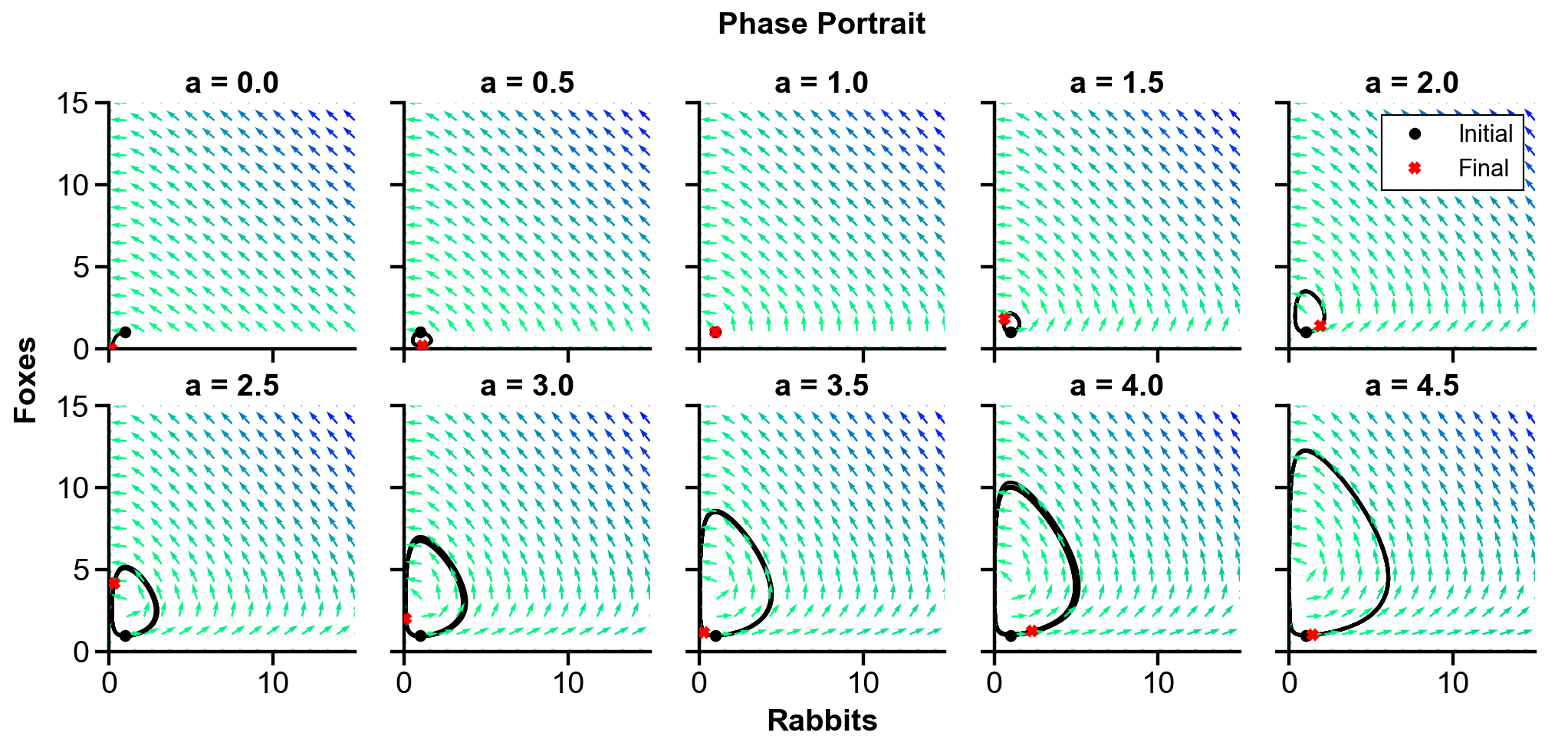
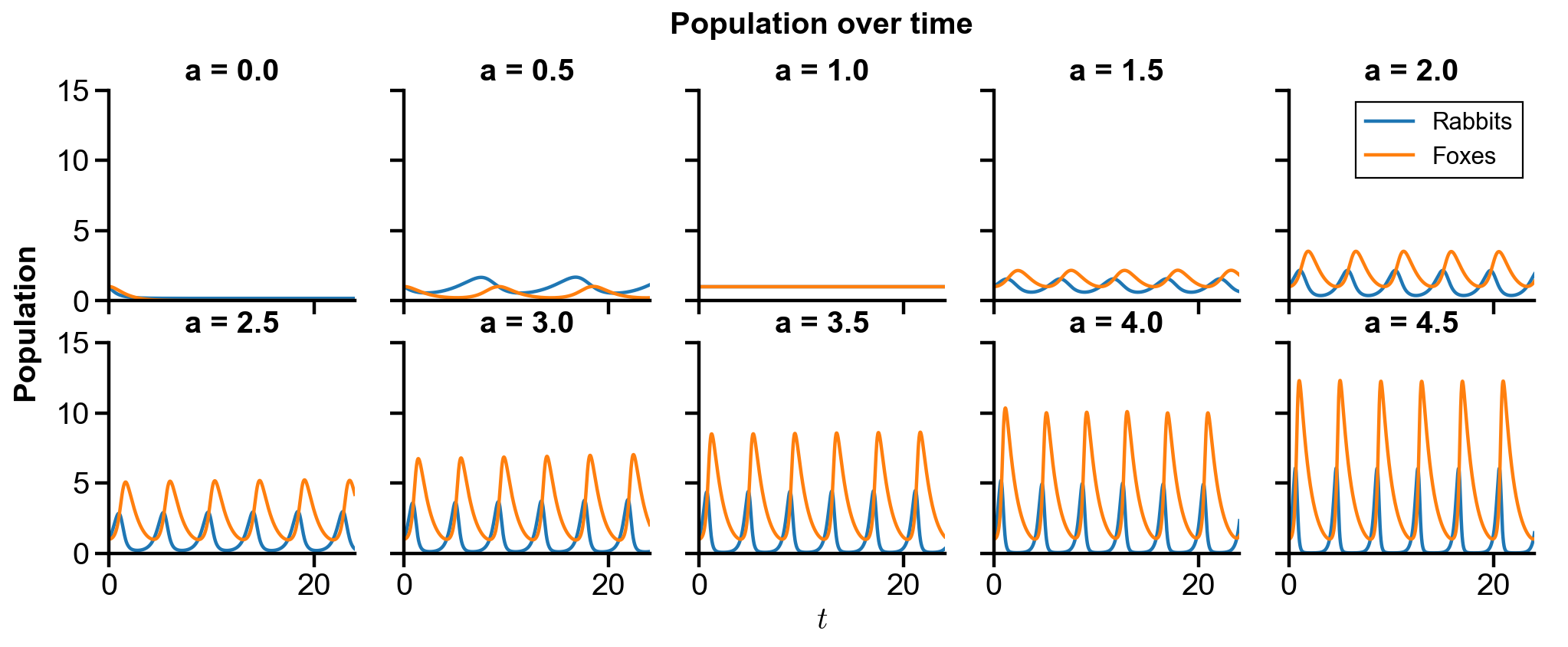
▲ The figure above shows the phase portrait of the rabbit-fox system and their population over time with changing
For
For
With the initial condition of
For larger
Changing
# model params
a = 1
b = np.arange(0.5, 5.5, 0.5)
c = 1
d = 1
# initial conditions
x0 = 1
y0 = 1
initial_val = np.array([x0, y0])
# time array
t_initial = 0
t_final = 12*2
t = np.linspace(t_initial, t_final, 1000)
# quiver grid
xvec = np.linspace(0, 15, 15)
yvec = np.linspace(0, 15, 15)
X, Y = np.meshgrid(xvec, yvec)
row = 2
col = 5
fig1, axs1 = plt.subplots(row, col, figsize=(col*3, row*3), sharex=True, sharey=True)
fig1.add_subplot(111, frameon=False)
plt.tick_params(labelcolor='none', which='both', top=False, bottom=False, left=False, right=False) # hide tick and tick label of the big axis
plt.xlabel('Rabbits')
plt.ylabel('Foxes')
plt.title('Phase Portrait', y=1.1)
fig2, axs2 = plt.subplots(row, col, figsize=(col*3, row*3-1), sharex=True, sharey=True)
fig2.add_subplot(111, frameon=False)
plt.tick_params(labelcolor='none', which='both', top=False, bottom=False, left=False, right=False) # hide tick and tick label of the big axis
plt.xlabel('$t$')
plt.ylabel('Population')
plt.title('Population over time', y=1.1)
for row_i in range(row):
for col_i in range(col):
i = row_i*col + col_i
# ode system
dxdt = lambda x, y : a*x - b[i]*x*y
dydt = lambda x, y : -c*y + d*x*y
ode_syst = lambda t, z : np.array([dxdt(*z), dydt(*z)])
# ode soln
ode_soln = scipy.integrate.solve_ivp(ode_syst, [t_initial, t_final], initial_val, t_eval=t)
# phase portrait
scale = np.sqrt(dxdt(X, Y)**2 + dydt(X, Y)**2)
axs1[row_i, col_i].quiver(X, Y, dxdt(X, Y)/scale, dydt(X, Y)/scale, scale, cmap='winter_r', scale=15, width=0.008) # regular
axs1[row_i, col_i].plot(*initial_val, 'o', color='black', label='Initial')
axs1[row_i, col_i].plot(ode_soln.y[0], ode_soln.y[1], color='black', zorder=0.5)
axs1[row_i, col_i].plot(ode_soln.y[0, -1], ode_soln.y[1, -1], 'X', color='red', zorder=4, label='Final')
# plot settings
axs1[row_i, col_i].set_xlim(0, 15)
axs1[row_i, col_i].set_ylim(0, 15)
axs1[row_i, col_i].set_title(f'b = {b[i] :.1f}')
axs1[row_i, col_i].set_aspect('equal')
axs2[row_i, col_i].plot(t, ode_soln.y[0], label='Rabbits')
axs2[row_i, col_i].plot(t, ode_soln.y[1], label='Foxes')
axs2[row_i, col_i].set_xlim(t_initial, t_final)
axs2[row_i, col_i].set_ylim(0, 15)
axs2[row_i, col_i].set_title(f'b = {b[i] :.1f}')
# compromised legend for common xylabels
if row_i == 0 and col_i == (col-1):
axs1[row_i, col_i].legend()
axs2[row_i, col_i].legend()
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
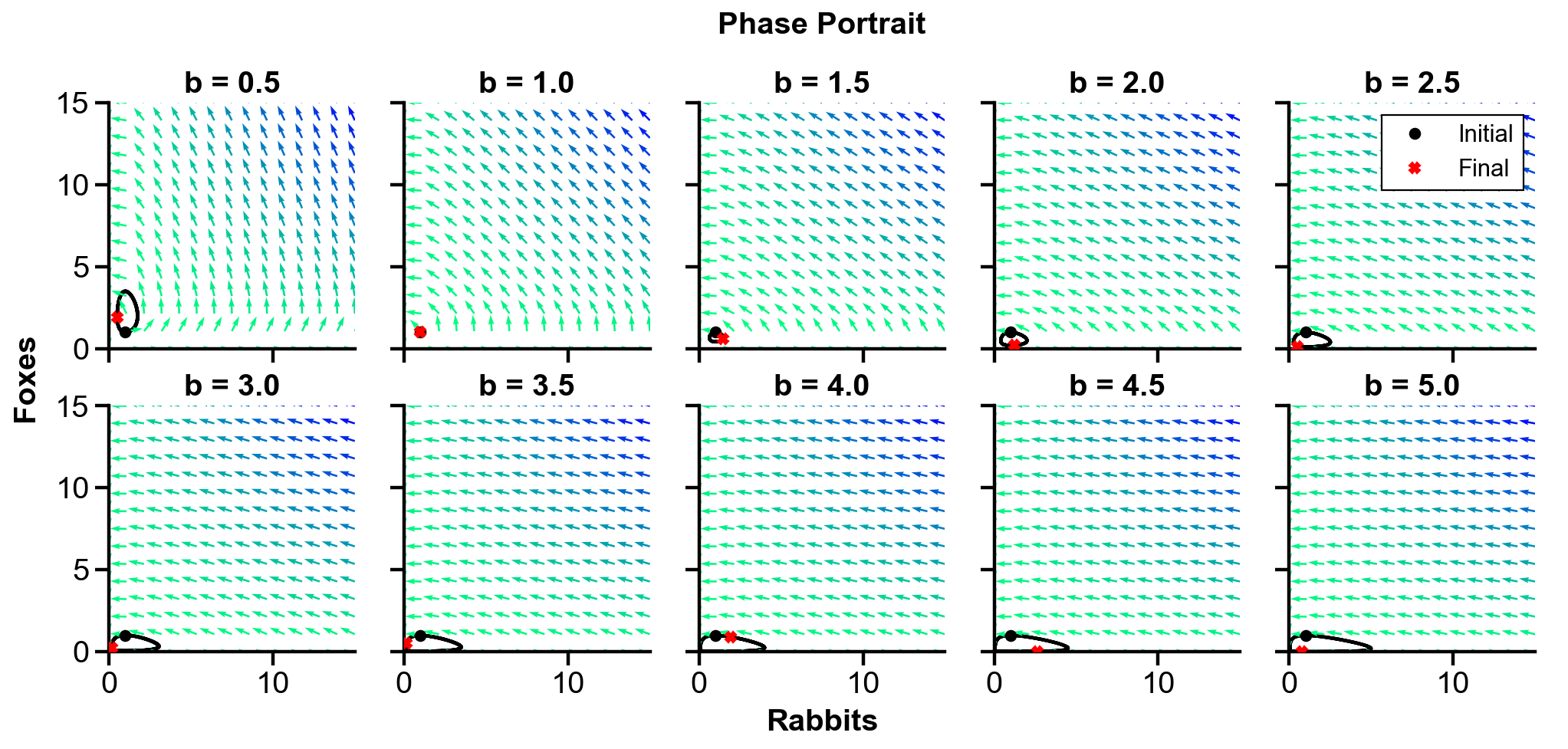
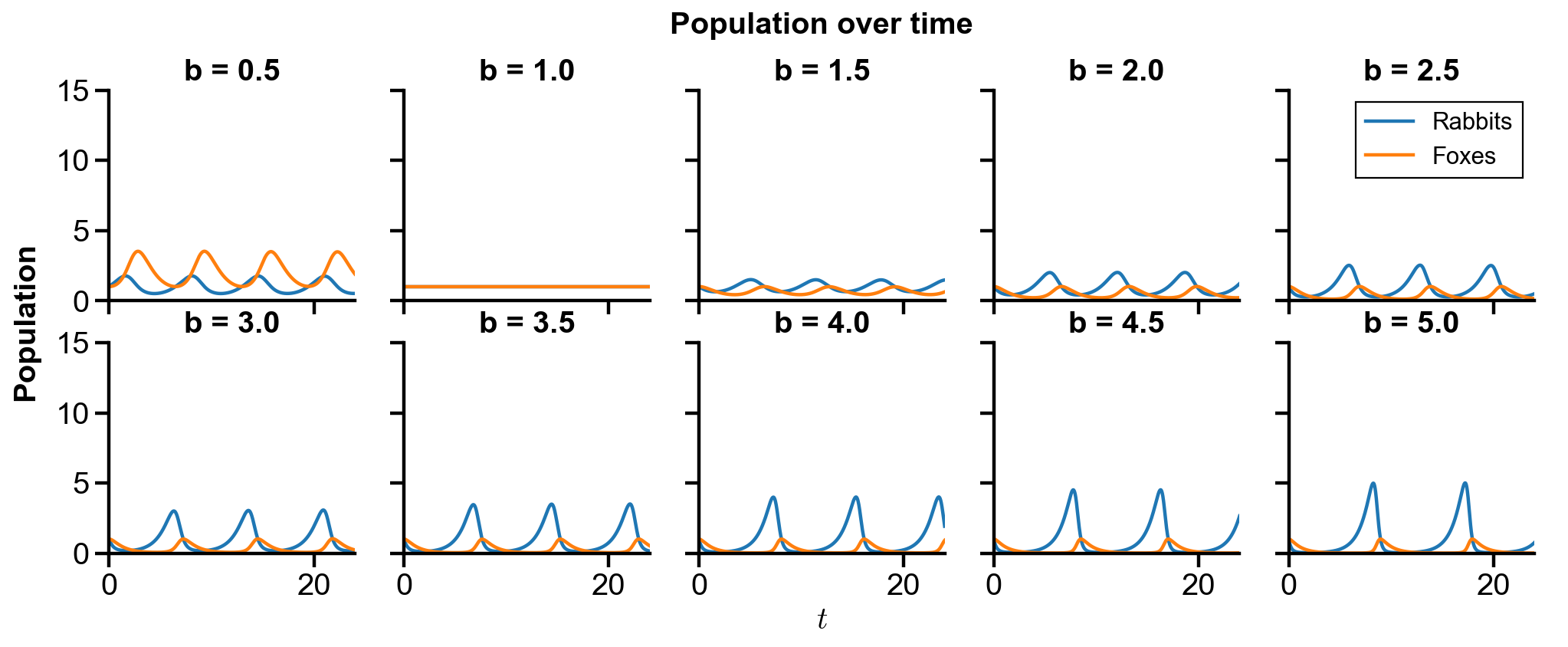
▲ The figure above shows the phase portrait of the rabbit-fox system and their population over time with changing
For
For
For
Changing
# model params
a = 1
b = 1
c = np.arange(0, 5, 0.5)
d = 1
# initial conditions
x0 = 1
y0 = 1
initial_val = np.array([x0, y0])
# time array
t_initial = 0
t_final = 12*2
t = np.linspace(t_initial, t_final, 1000)
# quiver grid
xvec = np.linspace(0, 15, 15)
yvec = np.linspace(0, 15, 15)
X, Y = np.meshgrid(xvec, yvec)
row = 2
col = 5
fig1, axs1 = plt.subplots(row, col, figsize=(col*3, row*3), sharex=True, sharey=True)
fig1.add_subplot(111, frameon=False)
plt.tick_params(labelcolor='none', which='both', top=False, bottom=False, left=False, right=False) # hide tick and tick label of the big axis
plt.xlabel('Rabbits')
plt.ylabel('Foxes')
plt.title('Phase Portrait', y=1.1)
fig2, axs2 = plt.subplots(row, col, figsize=(col*3, row*3-1), sharex=True, sharey=True)
fig2.add_subplot(111, frameon=False)
plt.tick_params(labelcolor='none', which='both', top=False, bottom=False, left=False, right=False) # hide tick and tick label of the big axis
plt.xlabel('$t$')
plt.ylabel('Population')
plt.title('Population over time', y=1.1)
for row_i in range(row):
for col_i in range(col):
i = row_i*col + col_i
# ode system
dxdt = lambda x, y : a*x - b*x*y
dydt = lambda x, y : -c[i]*y + d*x*y
ode_syst = lambda t, z : np.array([dxdt(*z), dydt(*z)])
# ode soln
ode_soln = scipy.integrate.solve_ivp(ode_syst, [t_initial, t_final], initial_val, t_eval=t)
# phase portrait
scale = np.sqrt(dxdt(X, Y)**2 + dydt(X, Y)**2)
axs1[row_i, col_i].quiver(X, Y, dxdt(X, Y)/scale, dydt(X, Y)/scale, scale, cmap='winter_r', scale=15, width=0.008) # regular
axs1[row_i, col_i].plot(*initial_val, 'o', color='black', label='Initial')
axs1[row_i, col_i].plot(ode_soln.y[0], ode_soln.y[1], color='black', zorder=0.5)
axs1[row_i, col_i].plot(ode_soln.y[0, -1], ode_soln.y[1, -1], 'X', color='red', zorder=4, label='Final')
# plot settings
axs1[row_i, col_i].set_xlim(0, 15)
axs1[row_i, col_i].set_ylim(0, 15)
axs1[row_i, col_i].set_title(f'c = {c[i] :.1f}')
axs1[row_i, col_i].set_aspect('equal')
axs2[row_i, col_i].plot(t, ode_soln.y[0], label='Rabbits')
axs2[row_i, col_i].plot(t, ode_soln.y[1], label='Foxes')
axs2[row_i, col_i].set_xlim(t_initial, t_final)
axs2[row_i, col_i].set_ylim(0, 15)
axs2[row_i, col_i].set_title(f'c = {c[i] :.1f}')
# compromised legend for common xylabels
if row_i == 0 and col_i == (col-1):
axs1[row_i, col_i].legend()
axs2[row_i, col_i].legend()
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
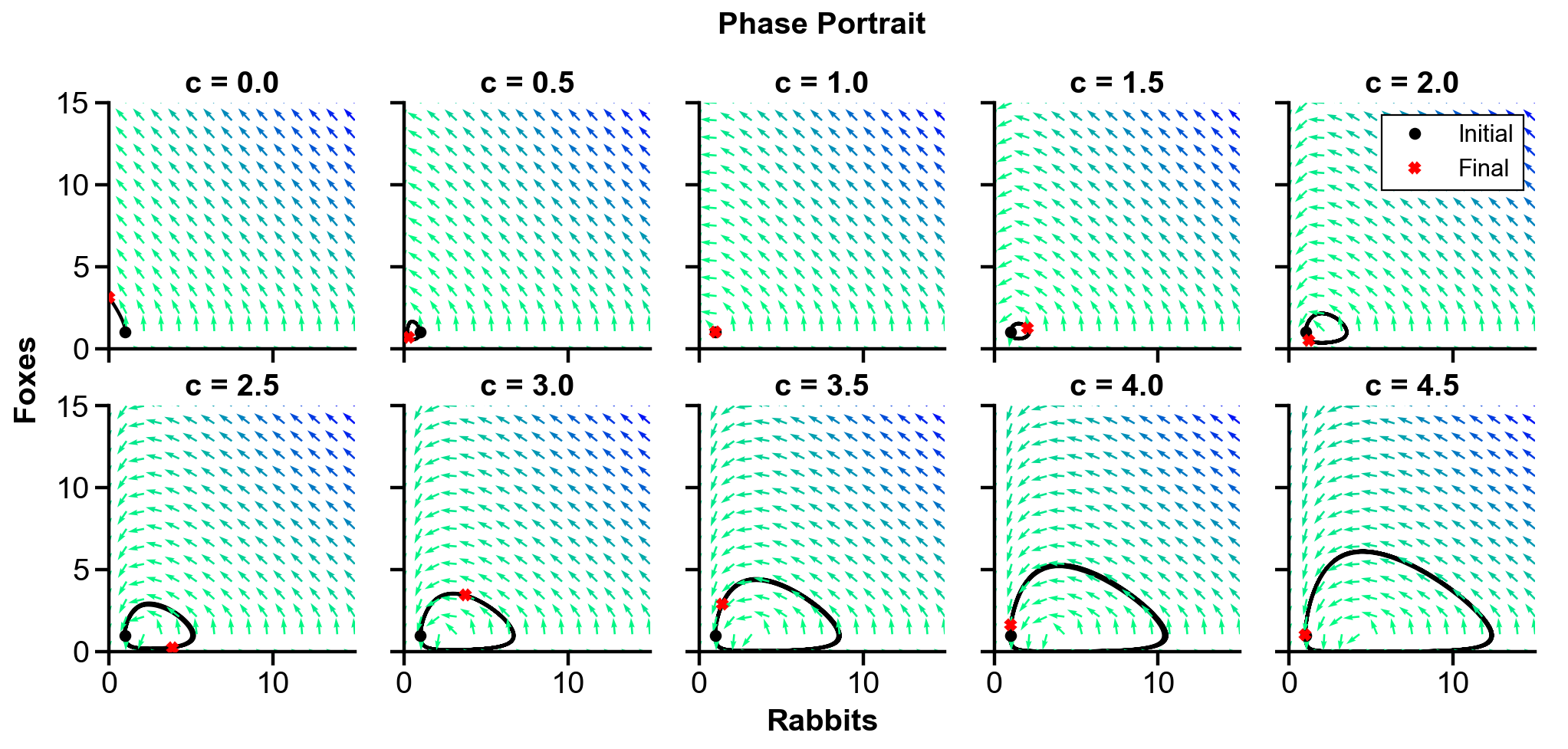
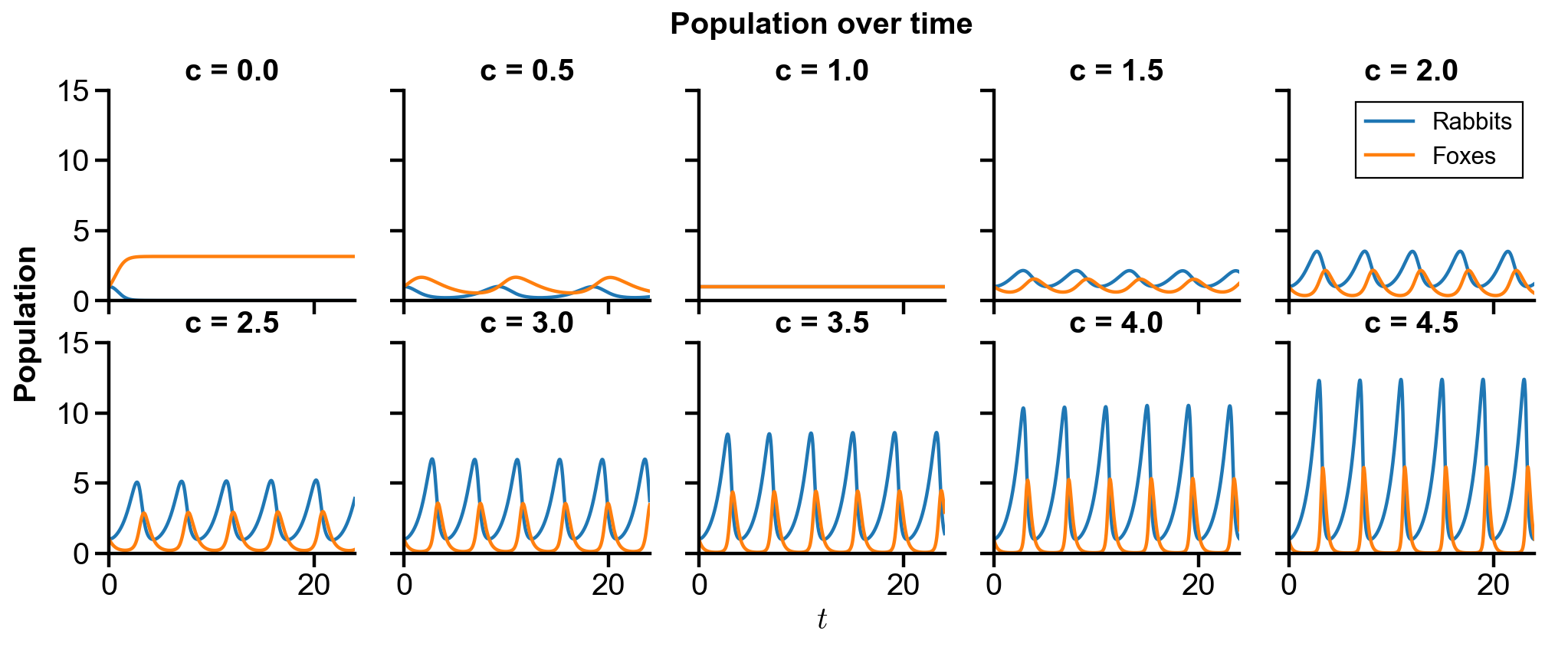
▲ The figure above shows the phase portrait of the rabbit-fox system and their population over time with changing
For
For
For
For
Changing
# model params
a = 1
b = 1
c = 1
d = np.arange(0, 5, 0.5)
# initial conditions
x0 = 1
y0 = 1
initial_val = np.array([x0, y0])
# time array
t_initial = 0
t_final = 12*2
t = np.linspace(t_initial, t_final, 1000)
# quiver grid
xvec = np.linspace(0, 15, 15)
yvec = np.linspace(0, 15, 15)
X, Y = np.meshgrid(xvec, yvec)
row = 2
col = 5
fig1, axs1 = plt.subplots(row, col, figsize=(col*3, row*3), sharex=True, sharey=True)
fig1.add_subplot(111, frameon=False)
plt.tick_params(labelcolor='none', which='both', top=False, bottom=False, left=False, right=False) # hide tick and tick label of the big axis
plt.xlabel('Rabbits')
plt.ylabel('Foxes')
plt.title('Phase Portrait', y=1.1)
fig2, axs2 = plt.subplots(row, col, figsize=(col*3, row*3-1), sharex=True, sharey=True)
fig2.add_subplot(111, frameon=False)
plt.tick_params(labelcolor='none', which='both', top=False, bottom=False, left=False, right=False) # hide tick and tick label of the big axis
plt.xlabel('$t$')
plt.ylabel('Population')
plt.title('Population over time', y=1.1)
for row_i in range(row):
for col_i in range(col):
i = row_i*col + col_i
# ode system
dxdt = lambda x, y : a*x - b*x*y
dydt = lambda x, y : -c*y + d[i]*x*y
ode_syst = lambda t, z : np.array([dxdt(*z), dydt(*z)])
# ode soln
ode_soln = scipy.integrate.solve_ivp(ode_syst, [t_initial, t_final], initial_val, t_eval=t)
# phase portrait
scale = np.sqrt(dxdt(X, Y)**2 + dydt(X, Y)**2)
axs1[row_i, col_i].quiver(X, Y, dxdt(X, Y)/scale, dydt(X, Y)/scale, scale, cmap='winter_r', scale=15, width=0.008) # regular
axs1[row_i, col_i].plot(*initial_val, 'o', color='black', label='Initial')
axs1[row_i, col_i].plot(ode_soln.y[0], ode_soln.y[1], color='black', zorder=0.5)
axs1[row_i, col_i].plot(ode_soln.y[0, -1], ode_soln.y[1, -1], 'X', color='red', zorder=4, label='Final')
# plot settings
axs1[row_i, col_i].set_xlim(0, 15)
axs1[row_i, col_i].set_ylim(0, 15)
axs1[row_i, col_i].set_title(f'd = {d[i] :.1f}')
axs1[row_i, col_i].set_aspect('equal')
axs2[row_i, col_i].plot(t, ode_soln.y[0], label='Rabbits')
axs2[row_i, col_i].plot(t, ode_soln.y[1], label='Foxes')
axs2[row_i, col_i].set_xlim(t_initial, t_final)
axs2[row_i, col_i].set_ylim(0, 15)
axs2[row_i, col_i].set_title(f'd = {d[i] :.1f}')
# compromised legend for common xylabels
if row_i == 0 and col_i == (col-1):
axs1[row_i, col_i].legend()
axs2[row_i, col_i].legend()
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:28: RuntimeWarning: invalid value encountered in true_divide
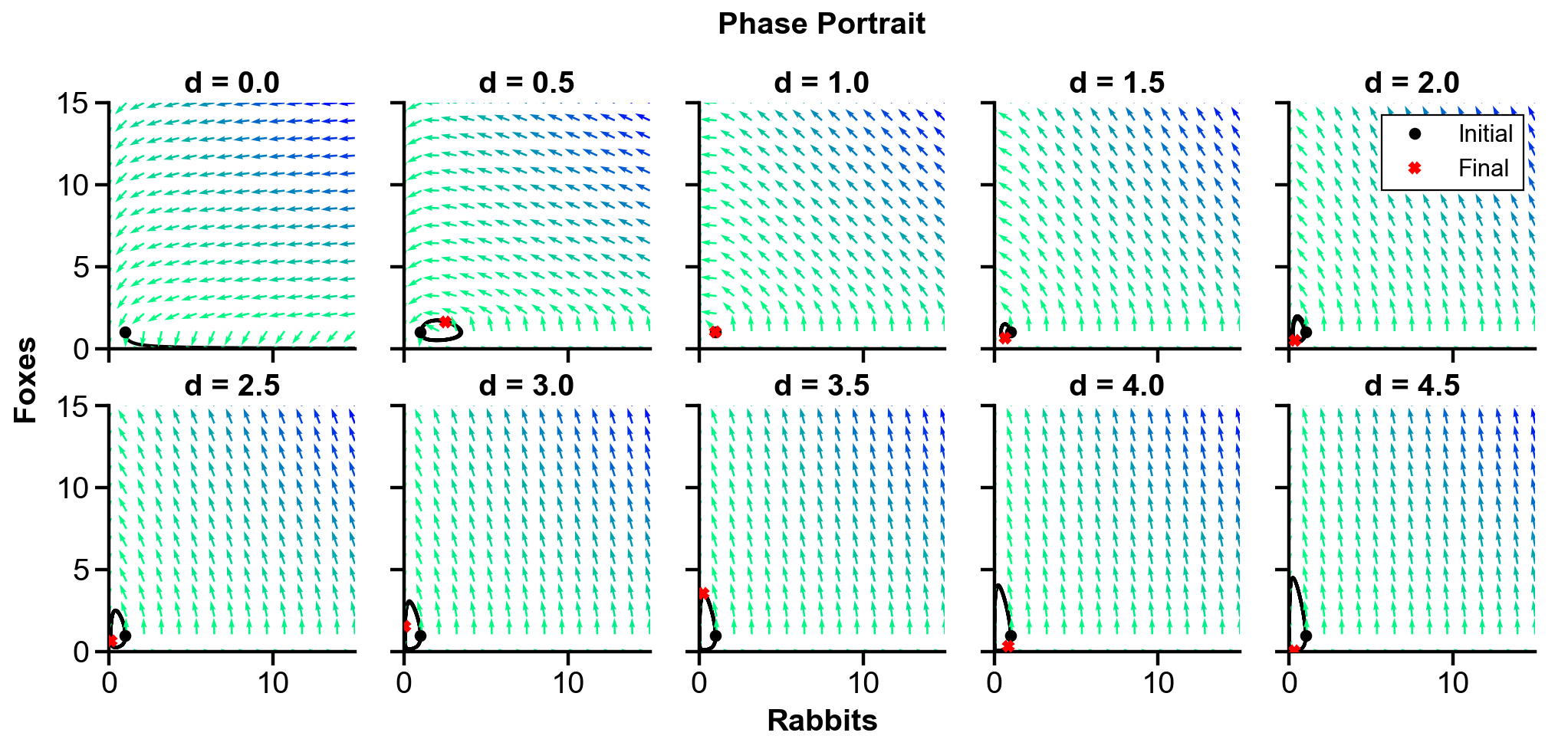
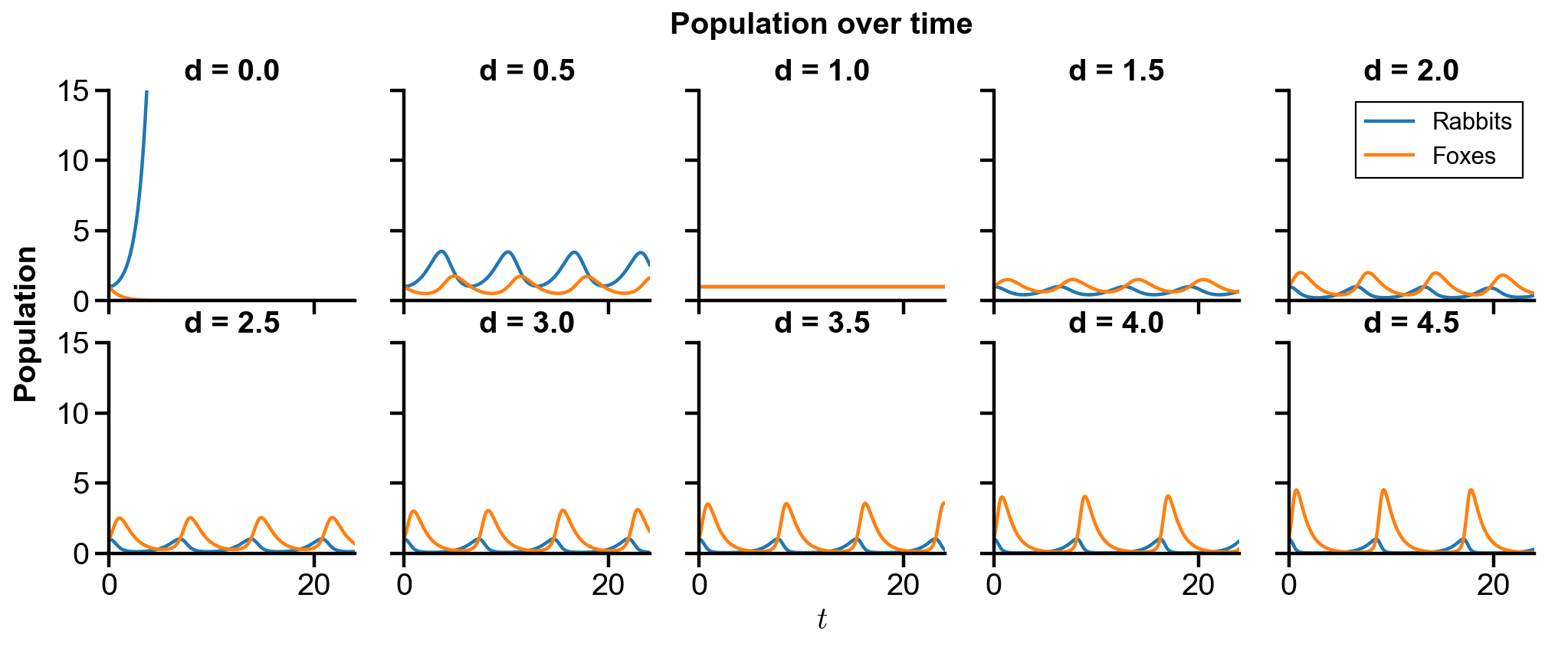
▲ The figure above shows the phase portrait of the rabbit-fox system and their population over time with changing
For
For
For
For
Acknowledgement#
I would like to thank Mr. Ben Trey, my high school everything teacher (Algebra II, AP Calculus BC, AP Physics 1, AP Chemistry, Applied Linear Algebra and Differential Equations), for introducing the concept of differential equation to me when I was in tenth grade (June 2017). We solved the Lotka-Volterra predator prey model with various parameters using the forward Euler method.
Here, we reproduce a subset of the result that was produced at that time.
Problem Statement. With the initial conditions of
Changing
Fixed other parameters
in
# model params
a = 10
b = 1
c = 1
d = np.arange(0.5, 5.1, 0.5)
# initial conditions
x0 = 10
y0 = 10
initial_val = np.array([x0, y0])
# time array
t_initial = 0
t_final = 12*2
t = np.linspace(t_initial, t_final, 1000)
# quiver grid
xvec = np.linspace(0, 15, 15)
yvec = np.linspace(0, 45, 15)
X, Y = np.meshgrid(xvec, yvec)
row = 2
col = 5
fig1, axs1 = plt.subplots(row, col, figsize=(col*3, row*3), sharex=True, sharey=True)
# share x and y labels
fig1.add_subplot(111, frameon=False)
plt.tick_params(labelcolor='none', which='both', top=False, bottom=False, left=False, right=False) # hide tick and tick label of the big axis
plt.xlabel('Rabbits')
plt.ylabel('Foxes')
plt.title('Phase Portrait', y=1.1)
fig2, axs2 = plt.subplots(row, col, figsize=(col*3, row*3-1), sharex=True, sharey=True)
# share x and y labels
fig2.add_subplot(111, frameon=False)
plt.tick_params(labelcolor='none', which='both', top=False, bottom=False, left=False, right=False) # hide tick and tick label of the big axis
plt.xlabel('$t$')
plt.ylabel('Population')
plt.title('Population over time', y=1.1)
for row_i in range(row):
for col_i in range(col):
i = row_i*col + col_i
# ode system
dxdt = lambda x, y : a*x - b*x*y
dydt = lambda x, y : -c*y + d[i]*x*y
ode_syst = lambda t, z : np.array([dxdt(*z), dydt(*z)])
# ode soln
ode_soln = scipy.integrate.solve_ivp(ode_syst, [t_initial, t_final], initial_val, t_eval=t)
# phase portrait
scale = np.sqrt(dxdt(X, Y)**2 + dydt(X, Y)**2)
axs1[row_i, col_i].quiver(X, Y, dxdt(X, Y)/scale, dydt(X, Y)/scale, scale, cmap='winter_r', scale=15, width=0.008) # regular
axs1[row_i, col_i].plot(*initial_val, 'o', color='black', label='Initial')
axs1[row_i, col_i].plot(ode_soln.y[0], ode_soln.y[1], color='black', zorder=0.5)
axs1[row_i, col_i].plot(ode_soln.y[0, -1], ode_soln.y[1, -1], 'X', color='red', zorder=4, label='Final')
# plot settings
axs1[row_i, col_i].set_xlim(0, 15)
axs1[row_i, col_i].set_ylim(0, 45)
axs1[row_i, col_i].set_title(f'd = {d[i] :.1f}')
axs2[row_i, col_i].plot(t, ode_soln.y[0], label='Rabbits')
axs2[row_i, col_i].plot(t, ode_soln.y[1], label='Foxes')
axs2[row_i, col_i].set_xlim(t_initial, t_final)
axs2[row_i, col_i].set_title(f'd = {d[i] :.1f}')
# compromised legend for common xylabels
if row_i == 0 and col_i == (col-1):
axs1[row_i, col_i].legend()
axs2[row_i, col_i].legend()
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:30: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:30: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:30: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:30: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:30: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:30: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:30: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:30: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:30: RuntimeWarning: invalid value encountered in true_divide
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:30: RuntimeWarning: invalid value encountered in true_divide
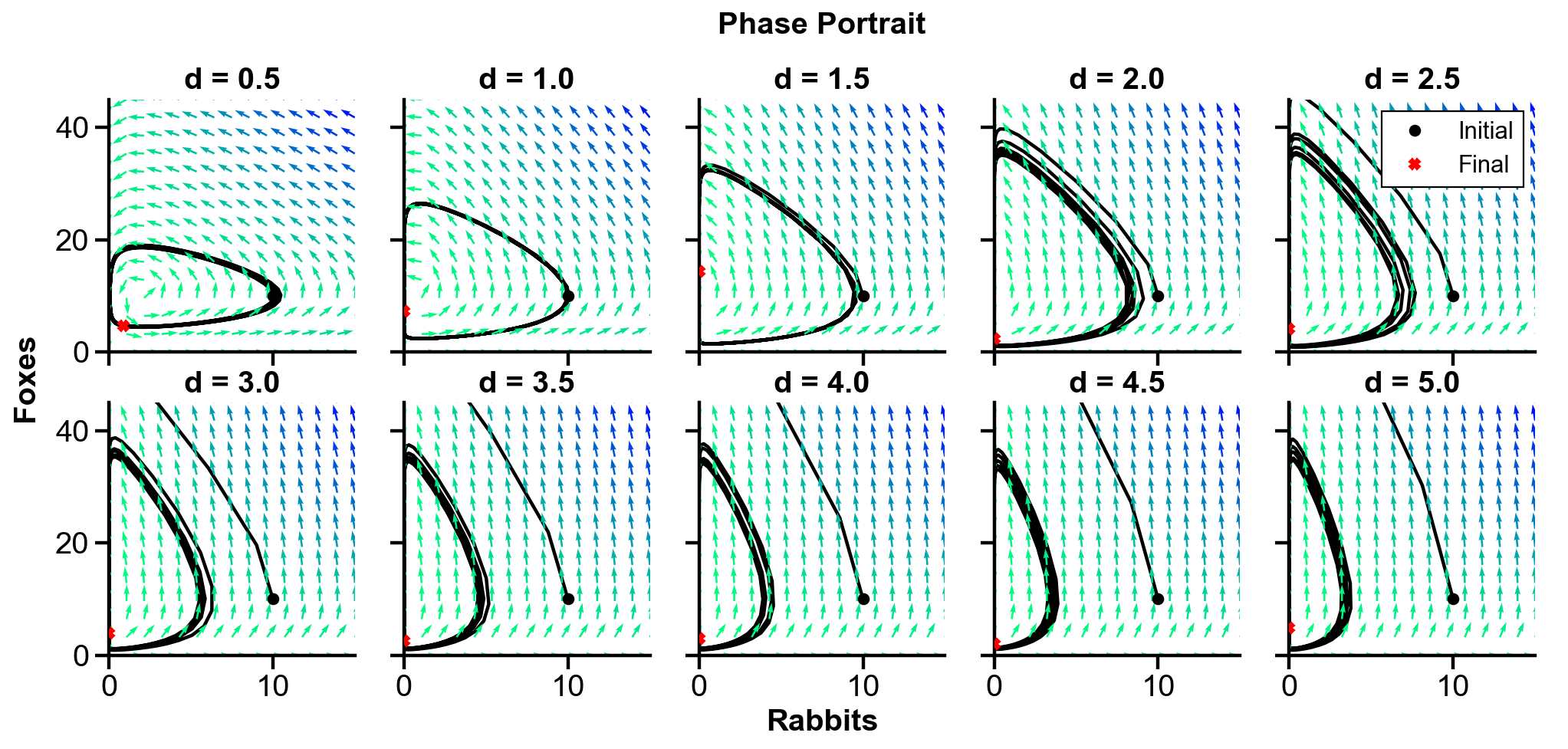
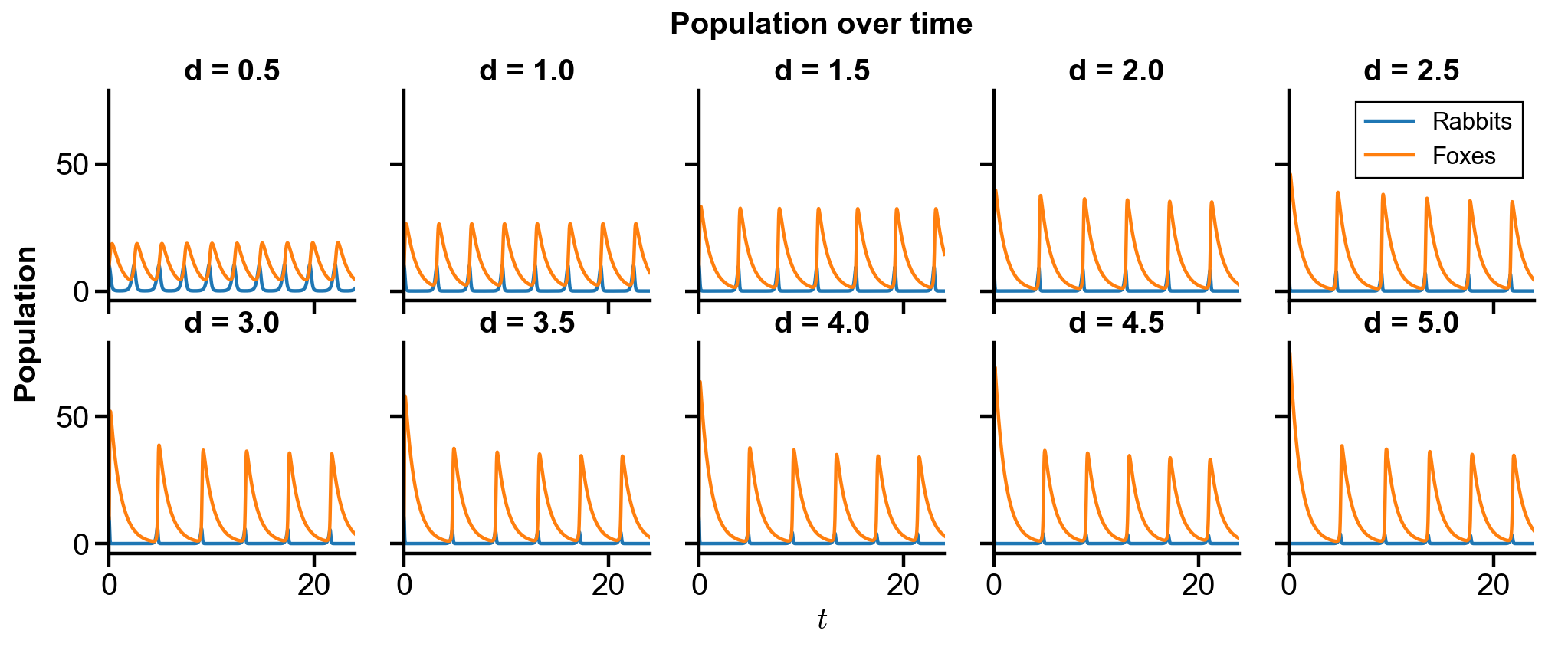
▲ Perfectly reproduced!
Competition model#
Let
Problem Statement. Solve the system of ODEs in
Plot the phase portrait and population over time.
Rabbit dominance#
# initial conditions
x0 = 1.5
y0 = 1
initial_val = np.array([x0, y0])
# time array
t_initial = 0
t_final = 10
t = np.linspace(t_initial, t_final, 1000)
# ode system
dxdt = lambda x, y : x*(3-x) - 2*x*y
dydt = lambda x, y : y*(2-y) - x*y
ode_syst = lambda t, z : np.array([dxdt(*z), dydt(*z)])
# ode soln
ode_soln = scipy.integrate.solve_ivp(ode_syst, [t_initial, t_final], initial_val, t_eval=t)
# quiver grid
xvec = np.linspace(0, 4, 25)
yvec = np.linspace(0, 4, 25)
X, Y = np.meshgrid(xvec, yvec)
fig, ax = plt.subplots(figsize=(4, 4))
# phase portrait
scale = np.sqrt(dxdt(X, Y)**2 + dydt(X, Y)**2)
ax.quiver(X, Y, dxdt(X, Y)/scale, dydt(X, Y)/scale, scale, cmap='winter_r', scale=20, width=0.005) # regular
ax.plot(*initial_val, 'o', color='black', label='Initial')
ax.plot(ode_soln.y[0], ode_soln.y[1], color='black', zorder=0.5)
ax.plot(ode_soln.y[0, -1], ode_soln.y[1, -1], 'X', color='red', zorder=4, label='Final')
# plot settings
ax.set_xlim(0, 4)
ax.set_ylim(0, 4)
ax.set_xlabel('Rabbits')
ax.set_ylabel('Sheep')
ax.set_title(f'Phase Portrait')
ax.set_aspect('equal')
ax.legend(loc='upper right')
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:4: RuntimeWarning: invalid value encountered in true_divide
after removing the cwd from sys.path.
<matplotlib.legend.Legend at 0x19ae60a4288>
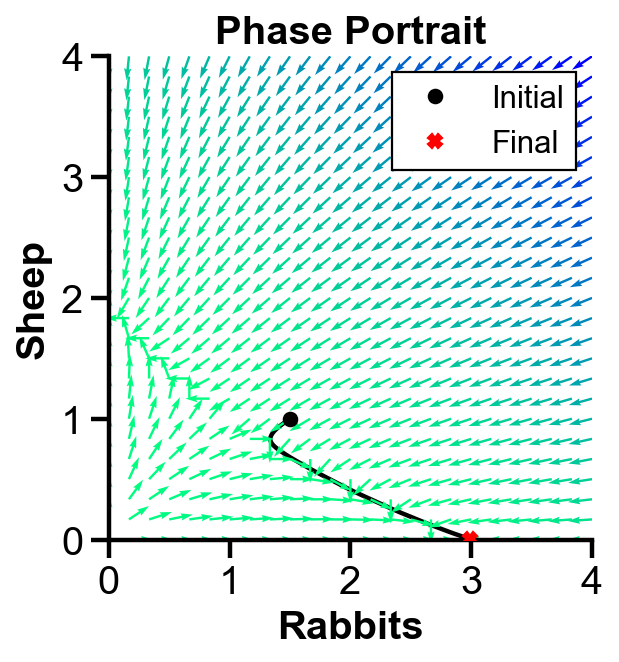
▲ The figure above shows the phase portrait with initial condition where rabbit population dominates sheep population for the system.
The population of both population first decreases. Then, the slope field pushes the rabbit population to increase and the sheep population to decrease. Sheep extincts over time.
fig, ax = plt.subplots(figsize=(4, 3))
ax.plot(t, ode_soln.y[0], label='Rabbits')
ax.plot(t, ode_soln.y[1], '--', label='Sheep')
# plot settings
ax.set_xlim(0)
ax.set_ylim(0)
ax.set_xlabel('$t$')
ax.set_ylabel('Population')
ax.set_title(f'Population over time')
ax.legend(loc='upper left', bbox_to_anchor=(1.05, 1.05))
<matplotlib.legend.Legend at 0x19ae7a87288>
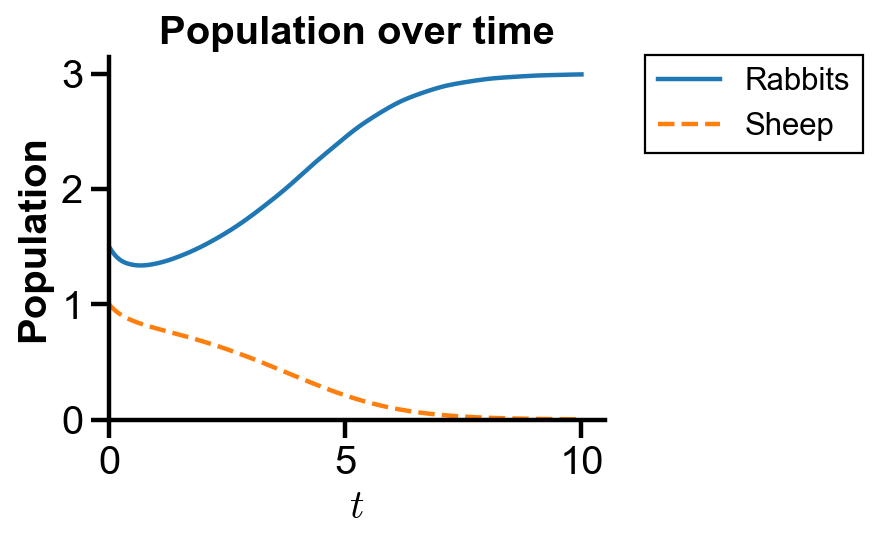
▲ The figure above shows the population over time with initial condition where rabbit population dominates sheep population for the system.
The figure confirms the initial decrease of both rabbit and sheep populations and the raise of rabbit population with the extinction of sheep.
Sheep dominance#
# initial conditions
x0 = 1
y0 = 1.5
initial_val = np.array([x0, y0])
# ode soln
ode_soln = scipy.integrate.solve_ivp(ode_syst, [t_initial, t_final], initial_val, t_eval=t)
fig, ax = plt.subplots(figsize=(4, 4))
# phase portrait
scale = np.sqrt(dxdt(X, Y)**2 + dydt(X, Y)**2)
ax.quiver(X, Y, dxdt(X, Y)/scale, dydt(X, Y)/scale, scale, cmap='winter_r', scale=20, width=0.005) # regular
ax.plot(*initial_val, 'o', color='black', label='Initial')
ax.plot(ode_soln.y[0], ode_soln.y[1], color='black', zorder=0.5)
ax.plot(ode_soln.y[0, -1], ode_soln.y[1, -1], 'X', color='red', zorder=4, label='Final')
# plot settings
ax.set_xlim(0, 4)
ax.set_ylim(0, 4)
ax.set_xlabel('Rabbits')
ax.set_ylabel('Sheep')
ax.set_title(f'Phase Portrait')
ax.set_aspect('equal')
ax.legend(loc='upper right')
C:\Softwares\Anaconda\Anaconda\lib\site-packages\ipykernel_launcher.py:4: RuntimeWarning: invalid value encountered in true_divide
after removing the cwd from sys.path.
<matplotlib.legend.Legend at 0x19ae608e608>
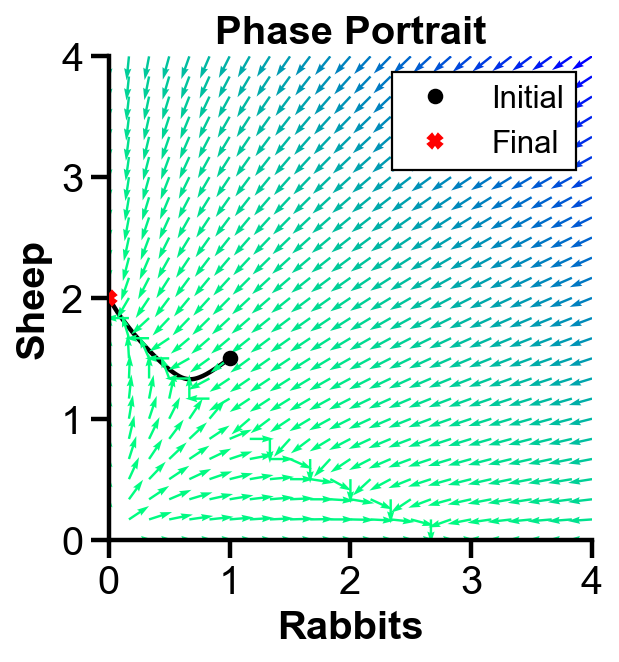
▲ The figure above shows the phase portrait with initial condition where sheep population dominates rabbit population for the system.
The population of both population first decreases. Then, the slope field pushes the sheep population to increase and the rabbit population to decrease. Rabbit extincts over time.
fig, ax = plt.subplots(figsize=(4, 3))
ax.plot(t, ode_soln.y[0], label='Rabbits')
ax.plot(t, ode_soln.y[1], '--', label='Sheep')
# plot settings
ax.set_xlim(0)
ax.set_ylim(0)
ax.set_xlabel('$t$')
ax.set_ylabel('Population')
ax.set_title(f'Population over time')
ax.legend(loc='upper left', bbox_to_anchor=(1.05, 1.05))
<matplotlib.legend.Legend at 0x19ae5cff088>
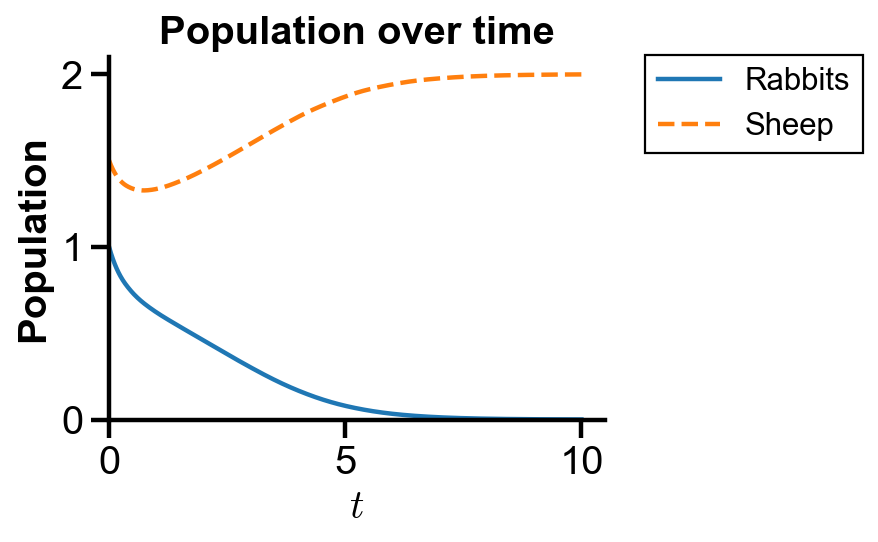
▲ The figure above shows the population over time with initial condition where sheep population dominates rabbit population for the system.
The figure confirms the initial decrease of both rabbit and sheep populations and the raise of sheep population with the extinction of rabbit.

